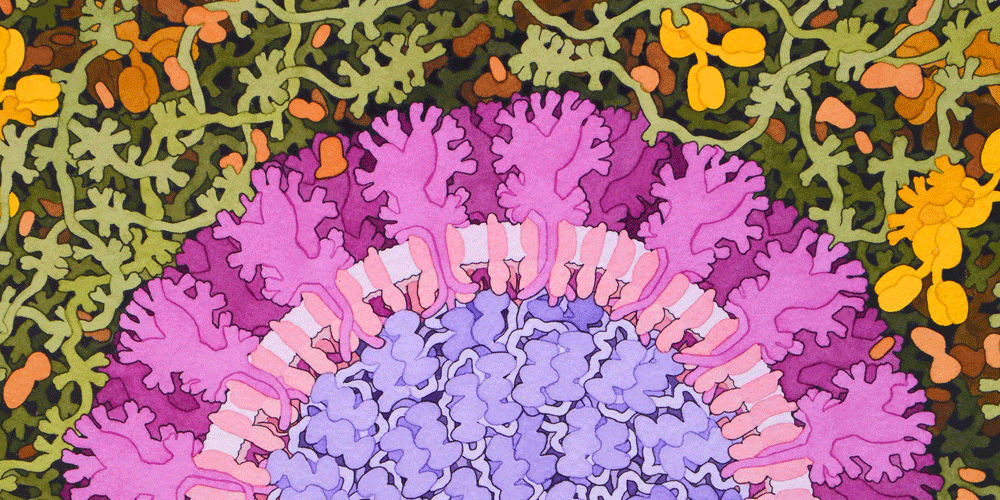
Knowledge sharing and public engagement are critical for finding solutions to the cascading crises caused by the SARS-CoV-2 pandemic. This article is part of an ISC blog series, which aims to highlight some of the latest COVID-19 related publications, initiatives and findings from ISC Members.
“The view is often defended that sciences should be built upon clear and sharply defined basal concepts. In actual fact, no science, not even the most exact, begins with such definitions. The true beginning of scientific activity consists rather in describing phenomena and then in proceeding to group, classify and correlate them.”
Freud S. (1915). “Instincts and their vicissitudes,” in: The Standard Edition of the Complete Psychological Works of Sigmund Freud (Vol. 14), Strachey J., editor. (London: The Hogarth Press).
Taxonomy – the discipline of classifying and naming things – is the bedrock of all the sciences. This was recognised by Sigmund Freud (1856—1939) as a psychological taxonomist, Dmitri Mendeleev (1834—1907) as a chemical taxonomist and Victor Goldschmidt (1888—1947) as a mineral taxonomist, and is recognised today by scientists at CERN as taxonomists of particle physics. However, for most people, taxonomy relates to biology – to organisms that reproduce and evolve. This taxonomy usually involves plants, animals and bacteria, but viruses are also in this category. Indeed, because of their spectacular diversity and abundance, viruses are both a challenge and an opportunity for taxonomists.
“Evolutionary mechanisms account for the unity and diversity of all species on Earth.”
Reece JB, et al. (2014). Chapter 1. Evolution, the Themes of Biology, and Scientific Inquiry. In Campbell Biology (10th ed.), Campbell N. et al., editors. (Boston: Pearson).
Virus taxonomy is important because it allows the clinical, biological and evolutionary features of a virus to be placed into a framework that accommodates and connects all viruses. The understanding that this brings has immense practical value. For example, when a virus emerges into humans from an animal reservoir, a taxonomic perspective puts us in a much better position to work out how it originated, which host it emerged from, how it replicates, how it causes disease, and how humans respond to being infected. As a result, we are much better placed to devise treatments and produce vaccines.
The novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is currently causing the COVID-19 pandemic, is a case in point. Taxonomy has helped us understand that the natural host of this virus is most likely to be bats and that its genetic material (genome) has evolved by the splicing together of different parental genomes (genetic recombination). Also, by knowing that the novel coronavirus is closely related to the SARS coronavirus that emerged in 2003, it has been possible to repurpose antiviral drugs such as remdesivir for the treatment of COVID-191 and to follow the lead that monoclonal antibodies generated from the cells of a patient infected with the 2003 coronavirus may be useful in the treatment of COVID-19 patients in 20202.
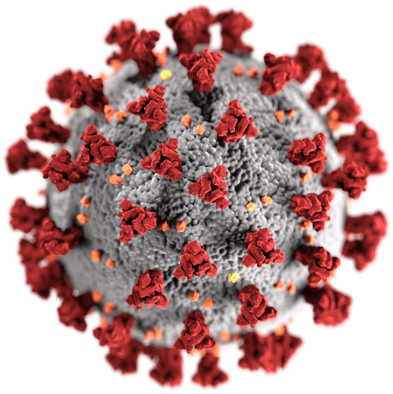
In December 2019, a novel coronavirus emerged in Wuhan, China. The virus is closely related to the 2003 SARS coronavirus and was, therefore, named severe acute respiratory syndrome coronavirus 2. SARS-CoV-2 is the causative agent of coronavirus infectious disease 2019 (COVID-19). By the end of June 2020, SARS-CoV-2 has infected more than 10 million people worldwide and there have been more than 500,000 COVID-19-associated deaths.
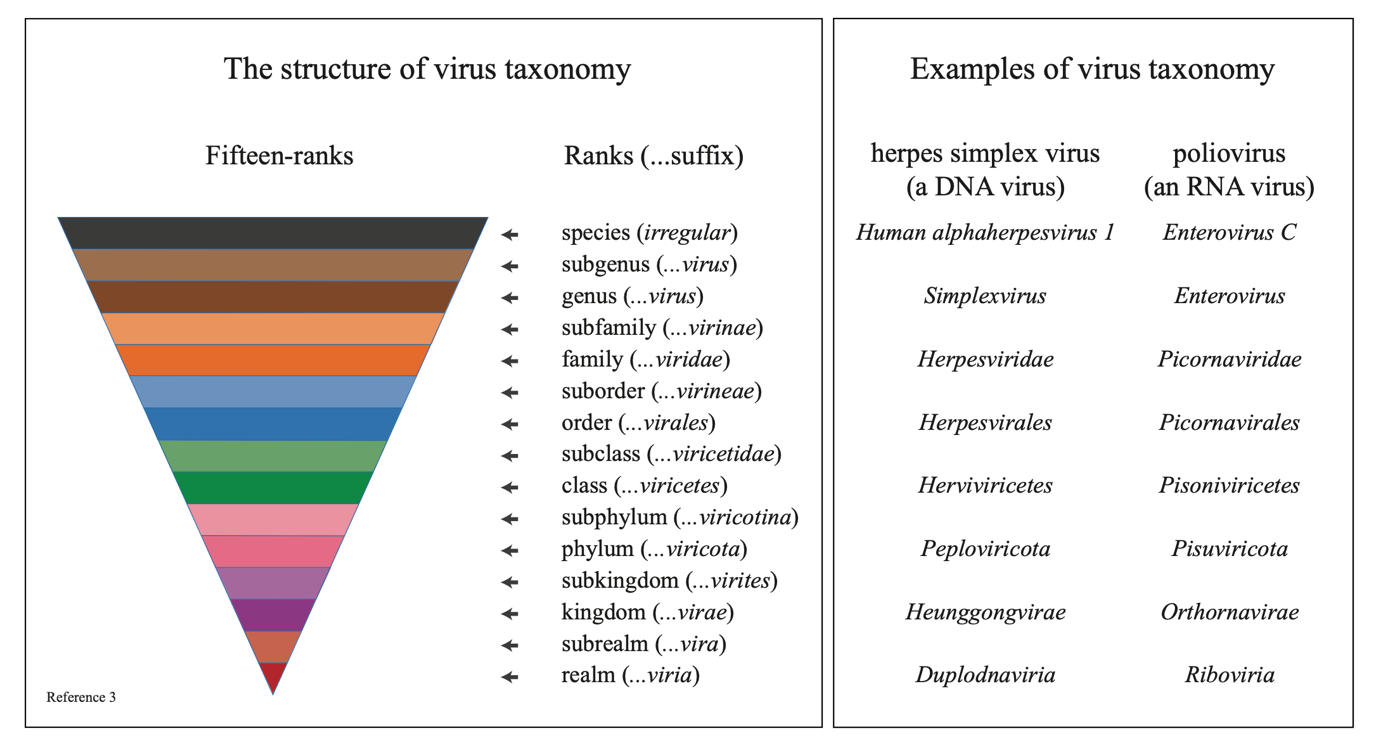
Like other organisms, viruses are classified into groups called taxa according to their similarities and dissimilarities. The lowest rank is the species. These are collected into genera, and so on up into families, orders, classes, phyla, kingdoms and realms. Overall, there is a hierarchical structure of 15 ranks that encompasses the full spectrum of virus diversity, stretching over an evolutionary timespan from when viruses first arose to the present day3.
Virus classification is traditionally a descriptive system based mainly on external (phenotypic) characters – the size and shape of a virus particle, the range of infected hosts and the diseases associated with infection. With the invention of ever more powerful DNA sequencing technologies, virus classification has gradually become a branch of evolutionary science. This change has accelerated in recent years because of the high-throughput sequencing of environmental samples (metagenomics), which has led to the detection of an astonishing range of viruses. Because of these advances, and the fact that we know nothing about these viruses except their genome, grouping viruses based on the relationships between their protein and nucleic acid sequences (phylogenetics) has become a key means of defining taxa.
There are still plenty of questions – for example, how do the taxa we create relate to the virus populations that we see in nature, and can we reliably reconstruct the early evolutionary history of viruses using only sequence information? Nonetheless, it is clear that we are entering a new era that will help us gain a much fuller understanding of the totality of viruses, their complex interactions with their hosts and their roles in natural ecosystems.
“Taxonomy is described sometimes as a science and sometimes as an art, but really it’s a battleground.”
Bryson B. (2003). In A Short History of Nearly Everything (New York City, The Broadway Press).
When combined with a good system for naming taxa, virus classification provides a powerful language of communication – in the classroom, in the laboratory, in the hospital, with the regulatory agencies, and with the public. The International Committee on Taxonomy of Viruses (ICTV) is responsible for developing this system. There are rules for naming taxa, in particular the endings used to denote the different ranks and the format in which names are written. The names of taxa other than species have been standardized for a long time, and the ICTV is now actively aiming to standardize species names to deal with the huge numbers of newly discovered viruses that will have to be classified and named in future. This nomenclature will most likely consist of two words (a binomial) – the name of the genus followed by a species epithet – as first described by Carl Linnaeus (1707—1778) almost 300 years ago.
One thing is for sure – virus taxonomy will go on exciting passions and intense debate!
In contrast to virus taxa, which are named by the ICTV, virus names are chosen by the experts who discover and research viruses. They are valid only in so far as they are accepted and used by the relevant community. There are no rules to govern this, but generally, viruses get their names from their hosts, the location in which they were first isolated, the manifestations of the diseases they cause, their phenotypic characters, or combinations of these things. It is surprising that despite the lack of regulation, virologists seem to work intuitively within an unwritten set of rules, using a limited number of naming patterns that enable the virus, the virus species and the disease to be distinguished from each other. Sometimes it works well – varicella-zoster virus belongs to the species Human alphaherpesvirus 3 and causes chickenpox (although it is not a “pox” virus). On occasion, the system is less successful – Japanese encephalitis virus belongs to the species Japanese encephalitis virus and causes Japanese encephalitis.
The naming of new virus diseases is also a rather haphazard process. When the virus infects humans and causes disease, the World Health Organization (WHO) recommends that disease names are based on the symptoms of the disease and, if the virus that causes the disease is known, it should be part of the name. On the other hand, the name should not include geographic locations, people’s names, cultural, population, industrial or occupational references, or terms that may incite undue fear. If these guidelines are not followed, the WHO itself may issue an interim name, which will normally be confirmed later in the International Classification of Diseases.
“Forewarned, forearmed, to be prepared is half the victory.”
Miguel de Cervantes Saavedra (1856). “Adventures of Don Quixote de la Mancha”. In Penguin Classics; Rev Ed edition, 2003, (London: Penguin Books).
Viruses have small genomes, short generation times and, at least in the case of RNA viruses, replication enzymes that are prone to making mistakes. Consequently, they evolve much more rapidly than other organisms. This provides unique opportunities to study the evolution of populations of circulating viruses (microevolution) as well as the evolution of viruses over much longer periods (macroevolution). Just as viruses have illuminated broad aspects of biology, for example by allowing us to dissect the workings of the cells they infect, they also provide an opportunity to road-test the taxonomic methods used throughout biology, including approaches to aligning sequences, predicting the functions of highly divergent proteins, and assessing the robustness of phylogenetic reconstruction programs involving large data sets. Future developments will improve the integration of the virus and cellular taxonomic systems and also have far-reaching impacts on the microbial, plant, veterinary, medical, computational and environmental sciences. Finally, it is obvious that we can no longer downplay the social, economic and personal impacts of the epidemics and pandemics that have been, and will continue to be, caused by viruses.
Stuart Siddell, ICTV Vice-President
Andrew Davison, ICTV President
www.ictv.global
1. Pruijssers AJ, George AS, Schäfer A, et al. Remdesivir potently inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Preprint. bioRxiv. 2020;2020.04.27.064279.
2. Pinto, D., Park, Y., Beltramello, M. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature (2020). https://doi.org/10.1038/s41586-020-2349-y
3. Gorbalenya, A.E., Krupovic, M., Mushegian, A. et al. The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat Microbiol 5, 668–674 (2020)
Photograph of Sigmund Freud: Max Halberstadt, 1921, Public Domain, Library of Congress.
Illustration of a coronavirus: 2020, Public Domain, Centers for Disease Control and Prevention, USA.
Examples of Virus Taxonomy. 2020, Copyright of the authors. May be used under a CC-BY-4.0 license, with acknowledgement to Reference 3.
Illustration by David S. Goodsell, RCSB Protein Data Bank; doi: 10.2210/rcsb_pdb/goodsell-gallery-019